
FDRdb overview
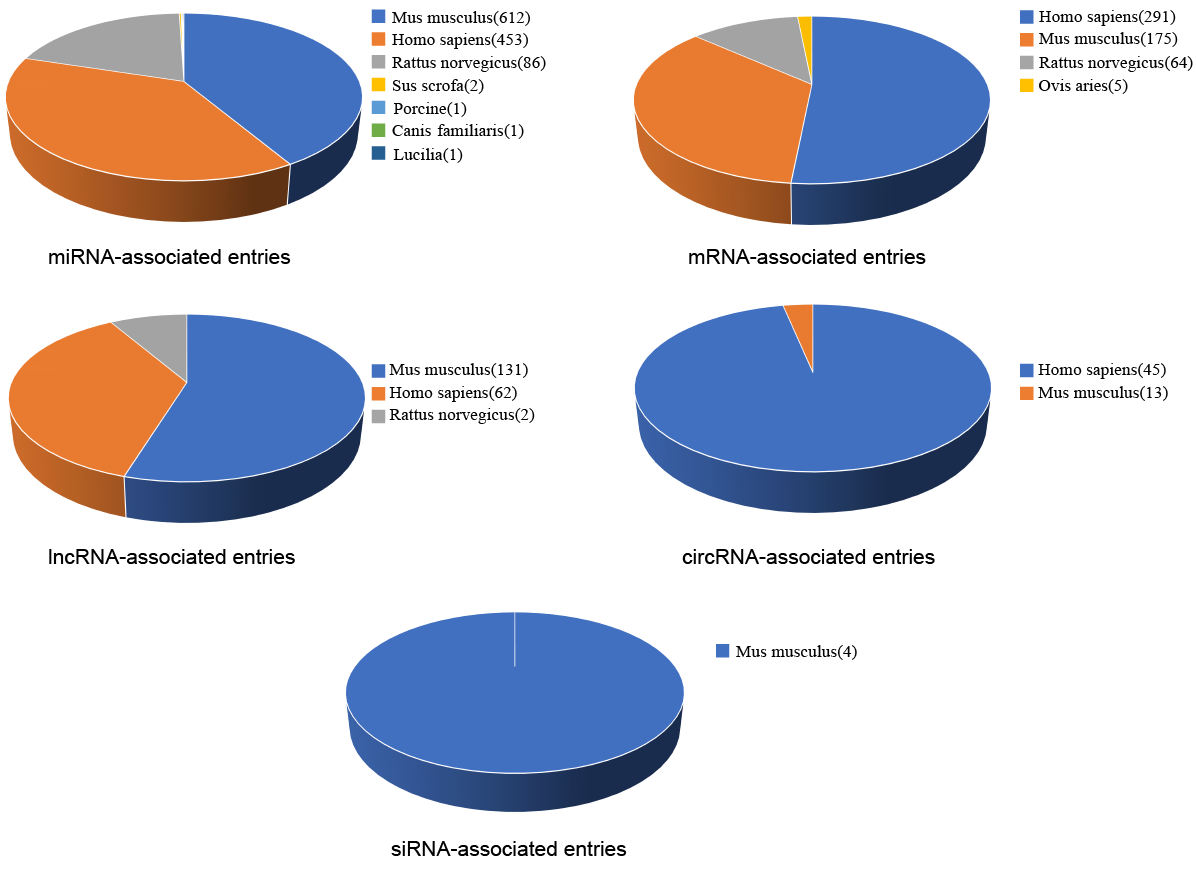
FDRdb is a powerful database that could help researchers to explore mechanisms of dysregulated RNAs in fibrotic diseases, search and analyze the fibrotic diseases-associated high-throughput datasets. The current version of FDRdb documents 1950 associations between 912 RNAs and 92 fibrotic diseases in 8 species, manually collected from 1127 published literatures. Meanwhile, FDRdb collects information of 765 genome datasets for fibrotic diseases.
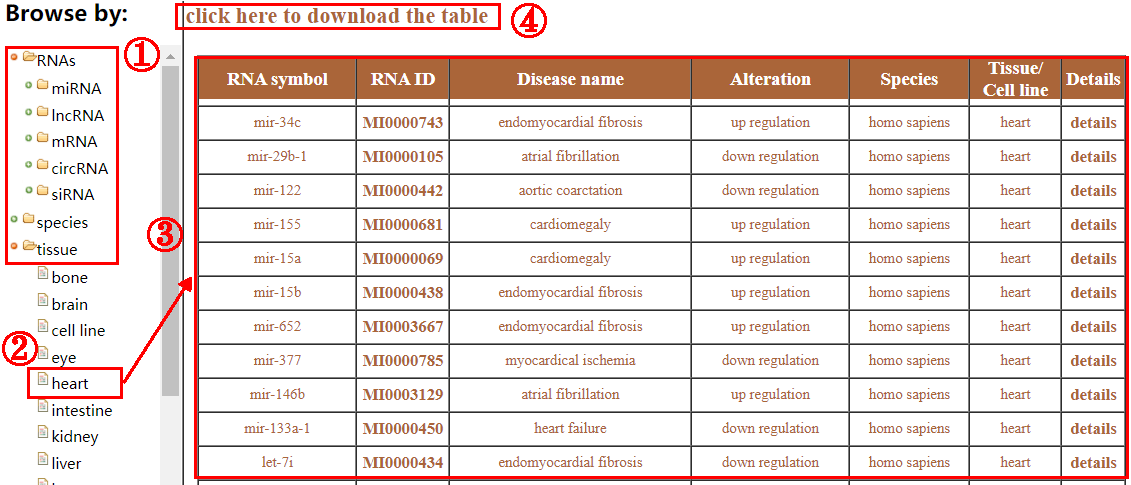
The browse page provides users with two options: browse by RNAs or datasets. On the Dataset browse page, the “optional tree” contains 4 categories: data type, species, disease and tissue. On the RNA browse page, the “optional tree” contains 5 types of RNAs (miRNA, lncRNA, mRNA, circRNA and siRNA), species, disease and tissue (1). Users can select the interested entries under the categories to view specific information about fibrosis (2-3). And, clicking on “click here to download” allows user to download the browsing table (4).
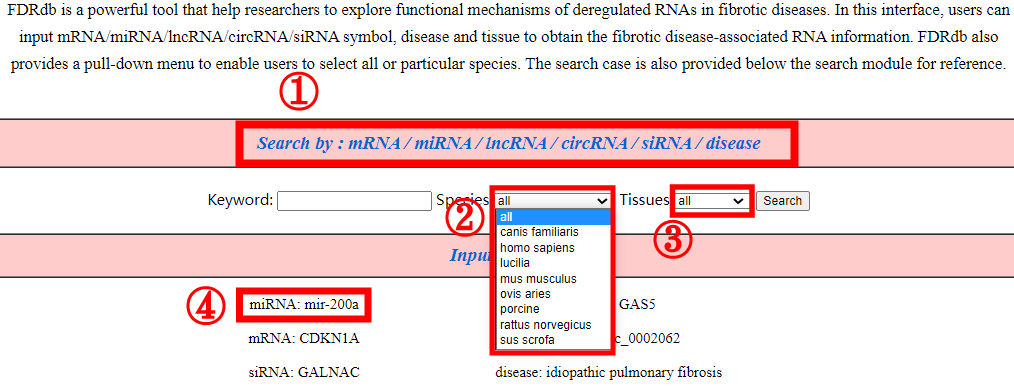
The search page allows users to enter keyword to search corresponding fibrosis-associated information. Users can search by different types of keywords: mRNA name, miRNA name, lncRNA name, circRNA name, siRNA name and fibrotic diseases (1). The search page provides users with pull-down menus to select all or specific species (2) or tissues (3). And, this page provides some searching examples on the bottom of the interface to help users to operate (4).
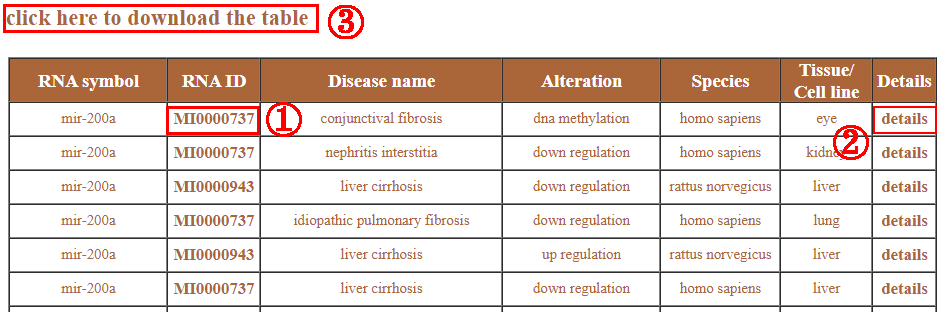
When users input keywords in the query box and press the “search” button, the searching results are showed in a table. Each row of the table contains one entry about your searching, and each column shows various types of information: RNA symbol, RNA ID, disease name, expression, species and tissue. The RNA ID links to the public database which contains details of this RNA (1). Users can click the “details” to get more detailed information (2). And clicking on “click here to download” allows the user to download the result table (3).
RNA Details
The RNA details page shows the specific information of your interested RNA. The information list contains: disease name, tissue, RNA name, RNA type, expression pattern, species, detection methods, RNA target, pathway, PubMed ID, reference title, publication time and fibrotic diseases-related function.

Data Details
The data details page shows specific information of the chosen dataset. The information list contains: disease name, tissue, the database of this dataset from, dataset ID, data type, data platform, species, samples number, PubMed ID, reference title, the brief description of the dataset, keyword of the dataset and the download link. Users can click “Download Link” button to jump to the original download page (1).
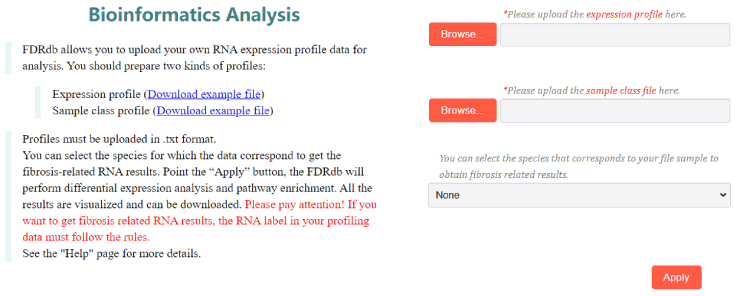
On the “Analysis” page, users can upload their own expression profile and sample classification data information to apply differential expression analysis and pathway enrichment. Users should upload an expression profile and a sample class file before analysis. In the expression profile, the first column is the sample labels, and the first columns is the RNA labels (please try to follow the following principles for the name of RNAs: mRNA labeled by symbol or entrez id; lncRNA named according to lncRNAdb or NONCODE rules; miRNA labeled by id in miRbase; circRNA named according to circBase. All data need to be preprocessed with log2. There are two rows in the sample class file: the first row is the sample labels, which is in the same order as the first line of the expression profile file; the second row is the class label of the corresponding samples. User can download example files, which are about idiopathic pulmonary fibrosis (IPF) (3). The two files must be uploaded in .txt format. Moreover, user can select specific species to get corresponding differential fibrotic RNAs between two types of samples (4). when users click “Apply” button, FDRdb will start to analyze your uploaded data and jump to the results page (5).
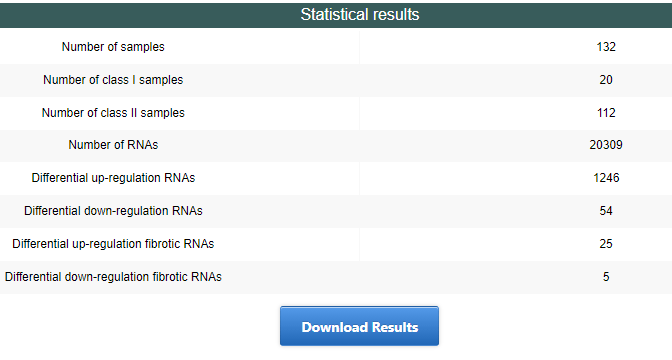
On the analysis result page, a brief summary of the results is listed on the top of the page. The result table statistics the differential RNA information between the two types of samples, and whether these differential RNAs are recorded in the FDRdb database as involved in fibrosis disease. All results and pictures can be downloaded on this page.
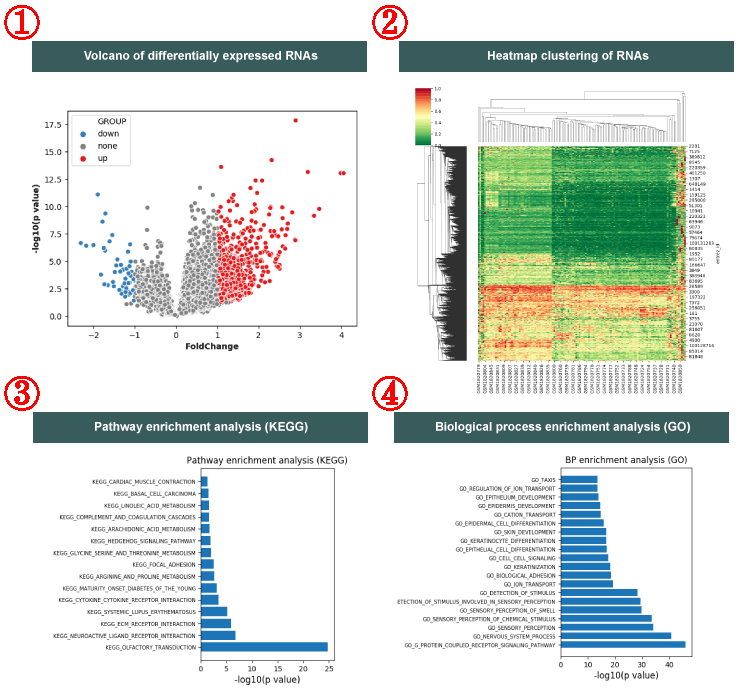
Meanwhile, the results are visualized as following: The volcano plot of differentially expressed RNAs (|log(fold-change)| > 1 and T-test, P < 0.05) (1). T the hierarchical clustering heatmap of differentially expressed RNAs (Hierarchical clustering) (2). Statistical bar chart of pathway enrichment analysis in KEGG (Hypergeometric test, P < 0.05) (3) and biological process enrichment analysis in GO (Hypergeometric test, P < 0.05) (4).
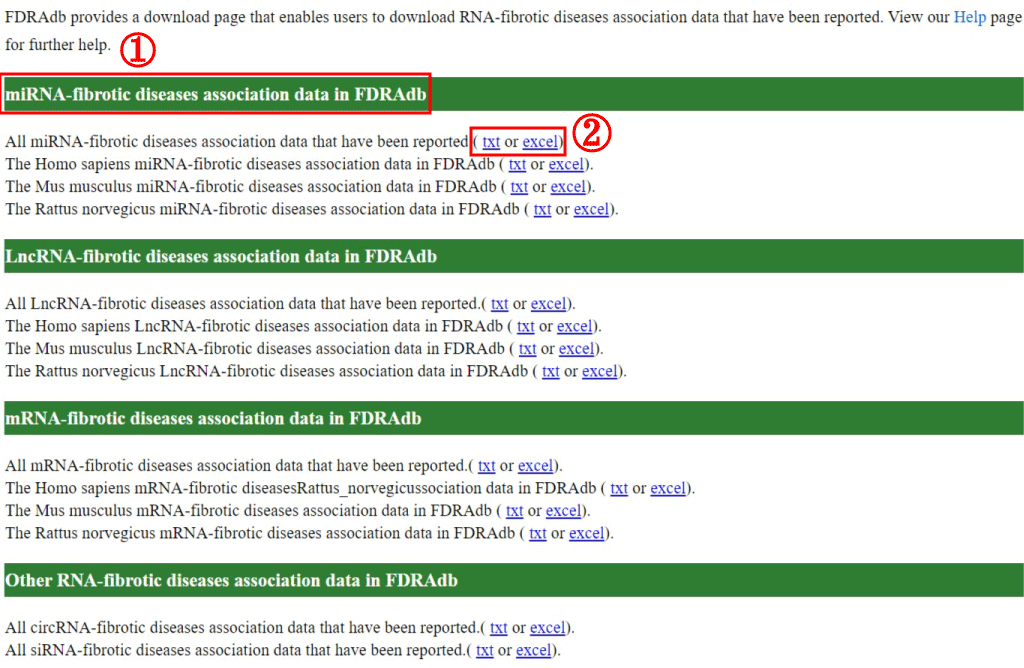
The download page allows users to download all RNA-fibrotic diseases relationship data in FDRdb (1). Users can download miRNA-fibrotic diseases, lncRNA-fibrotic diseases, mRNA-fibrotic diseases, circRNA-fibrotic diseases and siRNA-fibrotic diseases association data in .txt or .csv formats (2). FDRdb also allows users to download miRNA-fibrotic diseases, lncRNA-fibrotic diseases and mRNA-fibrotic diseases data by different species including Homo sapiens, Mus musculus and Rattus norvegicus.
-
April. 06, 2022
FDRdb update
The 423 new fibrosis-disease relationship and 332 new relevant datasets which published before 2022.04 were updated in FDRdb.
-
May. 04, 2021
The new model update
We added the bioinformatic analysis section into FDRdb.
-
March. 15, 2021
FDRdb
The original version of FDRdb were released in 2021.03.
Contact Information
Project Implementation
If you have any problems in using FDRdb,
or you just want to feedback to us, please contact us.
FDRdb: Fibrotic Disease-associated RNAome database
Contact lianghaihai@ems.hrbmu.edu.cn for inquiries and feedback
© All right reserved 2021