- 1.Overview Of EMT-Regulome
- 2.Search Menu
- 2.1 How To Search
- 2.2 Search Result--Infomation Box
- 2.3 Search Result--Motif List
- 2.4 Search Result--Network
- 3.Browse Menu
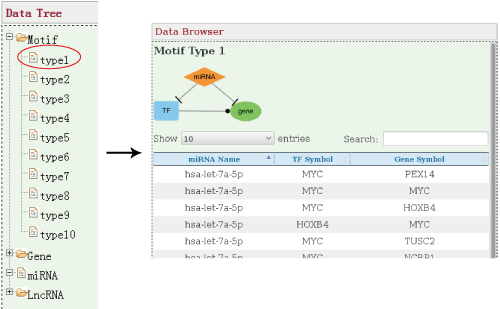
- 3.1 Motif
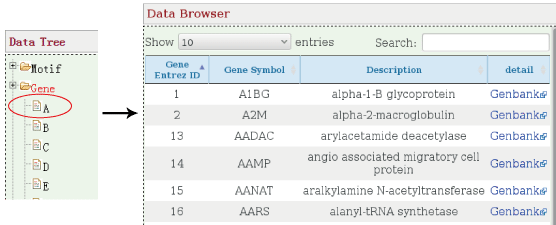
- 3.2 Nodes
- 4.Download Menu
- 4.1 Download Motif Data
The Epithelial-Mesenchymal Transition (EMT) is a highly conserved cellular process that allows polarized, immotile epithelial cells to convert to motile mesenchymal cells. EMT process governs embryonic development, tumor cell metastasis, organ fibrosis, tissue regeneration and so on. Transcription factors (TFs), such as Snail, Zeb and Twist, represses directly or indirectly the E-cadherin (CDH1) transcription. The inactivation of E-cadherin, which is considered as a hallmark of EMT. MicroRNA (miRNA) and long non-coding RNA (lncRNA) are also important regulators in EMT process. Those regulatory factors play co-regulation roles on the EMT process and the regulatory pattern acts as motifs, such as, feed-forward loops (FFLs) and competing endogenous RNA (ceRNA) relationship, which are the basic units in the EMT-related regulatory network. EMT-Regulome comprehensively collected the interactions between regulators and targets, including TF-target, miRNA-target, miRNA-lncRNA, TF-lncRNA and TF-miRNA and discerned ten types of co-regulation motifs for EMT process. EMT-Regulome provides the option of searching EMT-related regulatory relationships with motif and network.
In the “Search” page, the query function is composed of the basic search and other optional searches. For the coding gene, microRNA or the lncRNA, at least one search condition is required. To get a qualified search condition, the Entrez ID or the Symbol of genes are suggested and the Ensembl ID or Symbol of lncRNAs are acceptable. At present, the search condition of miRNAs only supports by miRNA name. Clicking the label of gene symbol "ZEB1" will present an example to search.
In addition, selecting types of motifs are available by ticking off corresponding box. The choice "ALL" is for the convenience of choosing all types. And in the “browse” page, there is an alternative way to search after choosing a specific file.

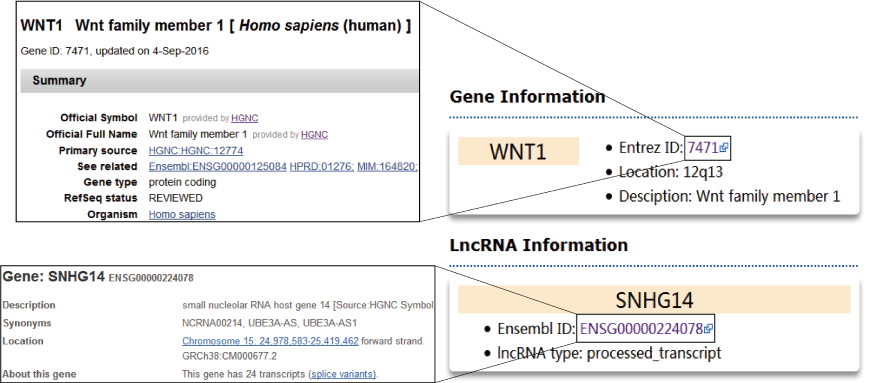
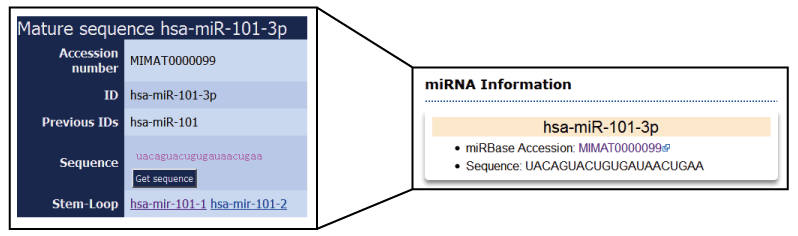
The information of the coding gene, miRNA or lncRNA, the key words used to be searched, will be listed in boxes at the top left corner of search result page. The box contains the gene symbol, Entrez ID with link to source website, the location in the chromosome and the brief description of the gene function. For a lncRNA, the box shows the Ensembl ID, symbol and type (lincRNA, antisense, sense intronic and so on) of lncRNA. For a miRNA, the box contains the accession number and mature sequence from miRBase.
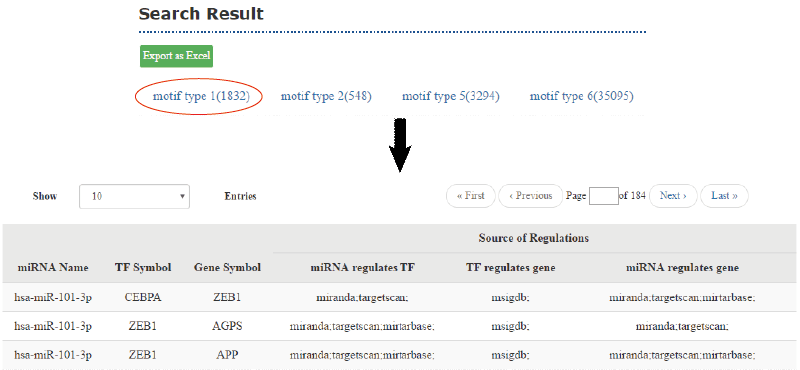
According to the motif types selected by the users, the returned motif list is showed. Once click out a tag, it will drop down and lists all results with name or symbol of each node in the motif and the regulation edge with source database information, which can be exported as excel files by click the button “Export as Excel”. Meanwhile, the corresponding network will be drawn on the right, with different layouts and can be exported in png format.
Networks consisting of more than 500 motifs will be time-consuming and induces the crash of web browser, thus we only display small-scale networks with motifs less than 500.
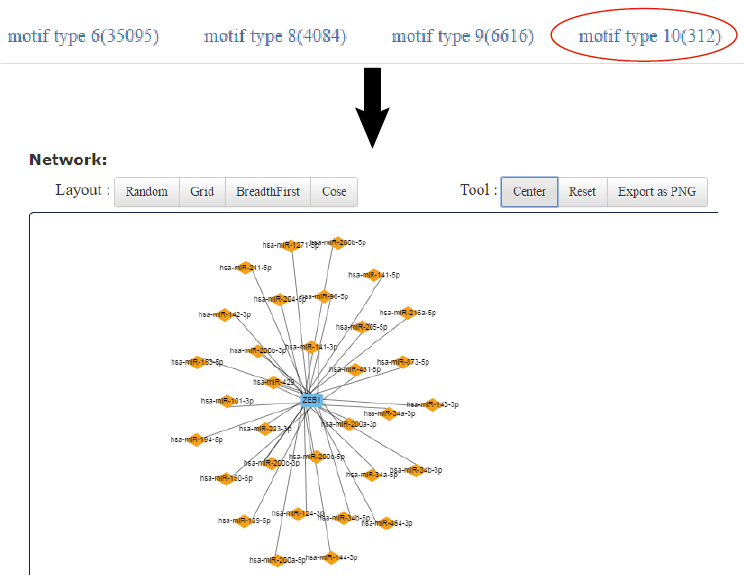
For the network shown on the right, legends on the top illustrate types of lines and nodes. T-shaped line represents the repression regulation. Lines with blunt arrow represents activated or repression regulation. Users are allowed to change the layout and move the nodes in the network.
The motifs are lined by the order of serial number and all motifs in a specific type will be displayed in the right window with pagination. Users can adapt how many records in each page. Some motif types like the ninth motif type with enormous amount need long time for waiting.
Information of genes, miRNAs and lncRNAs are offered in the page. Coding genes and lncRNAs are sorted by the order of letters and miRNAs are recruited in a file. Their corresponding external links of concrete information are provided respectively, including NCBI, miRBase and Ensembl.
All types of motifs in the EMT-Regulome are provided with download links in "Download" page. Users can conveniently access the link by clicking the navigation on the right.
Yang et al.(Integrated analyses identify a master microRNA regulatory network for the mesenchymal subtype in serous ovarian cancer. Cancer cell 2013;23(2):186-199) have classified the TCGA ovarian cancer samples into integrated epithelial (iE) subtype and integrated mesenchymal (iM) subtype labels. Using these labels we defined activated motifs as the nodes in motifs were differentially expressed in mesenchymal type compared with the epithelial type (T-test with P<0.05, 0.01 or FDR<0.1) and the edges in motifs were differentially co-expressed in mesenchymal type (Pearson Correlation test with P<0.05).