INTRODUCTION:
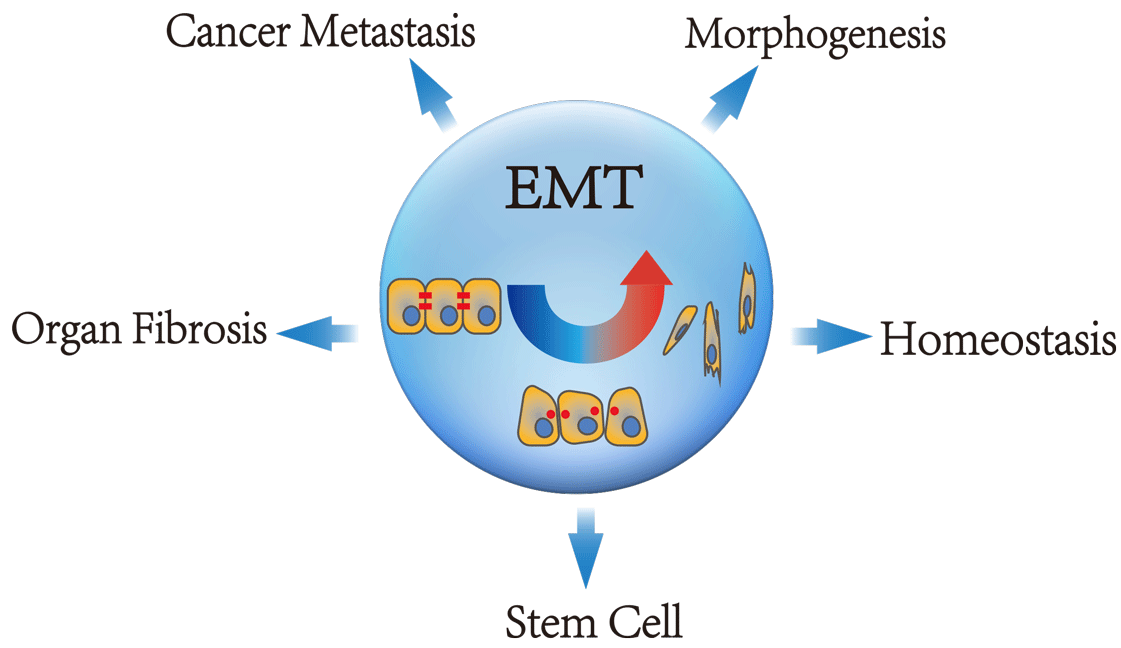
The Epithelial-Mesenchymal Transition (EMT) allows polarized, immotile epithelial cells to transform into motile mesenchymal cells, which is a fundamental cellular process that governs embryonic development, tumor cell metastasis, organ fibrosis, tissue regeneration and so on. EMT is regulated by many signaling pathways.The TGF-b signaling pathway has an established role in promoting EMT by down-regulating E-cadherin via transcription factors (TFs), such as Twist, Snail and Slug. Multiple TF-target relationships form a regulatory transcriptional network. Moreover, the post-transcriptional regulatory network consisting of microRNA(miRNA) and long non-coding RNA(lncRNA) also dominates the process of EMT. Comprehensively investigation of co-regulations of regulators(TF, miRNA and lncRNA) on EMT could speed up the elucidation of the potential mechanism of EMT in development, fibrosis and cancer progression. EMT-Regulome database systematically collects the regulations among TF, miRNA, lncRNA and EMT genes and records diverse regulatory motifs involved in EMT process.
MOTIFS TYPE
| No. | Type |
Legend:

LINKS TO OTHER DATABASES
- TargetScan
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 2015;4
- miRBase
miRBase: microRNA sequences, targets and gene nomenclature.
Nucleic Acids Res,2006;34(Database Issue):D140-D144
- miRTarBase
miRTarBase: a database curates experimentally validated microRNA-target interaction.
Nucleic Acids Res,2011;39(Database Issue):D163-9
- TRANSFAC
TRANSFAC: transcriptional regulation, from patterns to profiles.
Nucleic Acids Res,2003;31:374-8
- miRanda
The microRNA.org resource: targets and expression.
Nucleic Acids Res,2008;36(Database Issue):D149-53
- dbEMT
dbEMT: an epithelial-mesenchymal transition associated gene resource.
Scientific Reports,2015;5:11459
- miRcode
miRcode: a map of putative microRNA target sites in the long non-coding transciptome.
Bioinformatics 2012;28(15):2062-3
- starBase v2.0
starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data.
Nucleic Acids Res,2014;42(Database Issue):D92-7
- MsigDB
Molecular signatures database(MSigDB) 3.0.
Bioinformatics,2011;27(12):1739-40
- ChIPBase
ChIPBase: a database for decoding the thanscriptional regulation of long non-coding RNA and microRNA genes from ChIP-Seq data.
Nucleic Acids Res,2013;41(Database Issue):D177-87
- Transmir
TransmiR: a transcription factor-microRNA regulation database.
Nucleic Acids Res,2010;38(Database Issue):D119-22